The National Cancer Institute Real Time Picture Processor
| Anchor | ||||
|---|---|---|---|---|
|
...
Many properties of biological materials can be visualized directly using microscopy, electrophoresis, or other visualization mechanisms. The image subjects may have been improved before digital image capture using various detection-enhancement methods (such as stains, dyes, autoradiography, phase-contrast, interference microscopy, etc.) to visualize the data of interest. Digital image processing (see wikipedia.org and and dictionary.com entries) is a method for the separation, detection, and quantification of the objects of interest in biological materials. Quantified data helps scientists perform more rigorous analyses of their biological experiments and improve the conclusions of their analyses.
There are two major goals of this history: to document the events and conditions that led to the creation of one of the first grayscale image processors, and to describe the highly effective complementary collaboration that allowed alloAsilomar Third Engineering Foundation Conference on Automated Cytologywed this project to flourish. Occasionally, references will be made to other later advances indirectly related to the RTPP work that would not have happened without the RTPP. Where possible, we have linked to open access journal PDFs, and have included PDFs of the key technical reports describing the RTPP on this Web site.
...
The RTPP project was conceived and initiated by Dr. Lewis "Lew" Lipkin, M.D., head of the Image Processing Unit, later the Image Processing Section (IPS), in the the National Cancer Institute (NCI). The intellectual concept behind computer-controlled microscopy started in 1962 when Lew was an assistant professor of neuropathology at Downstate Medical Center in New York. Professor Patrick Fitzgerald, Chairman of the Pathology Department at Downstate, was studying pancreatic cell growth. Dr. Vinichaichol, who was doing visual grain counts on thin pancreatic sections, was finding mixed results. The problem was statistical. Dr. Lipkin was asked to design a proper sampling technique. Grain counting was Grain counting was a method used to measure cell metabolism before the days of antibody techniques applied to living cells and fluorescent techniques that came about during our time in NIH. Lew, who happened to know something about statistics, was asked by Dr. Fitzgerald to find out what was wrong with his statistics. After some thought, Lew realized that Dr. Vinichaichol was staying in one area of the slide and he had no way of knowing when he was recounting the same cells. Lew didn't want to continue looking at biological material that he couldn't explore without using some form of quantification.
...
Dr. Richard Masland, M.D., the director of the National Institute of Neurological Disease and Blindness (NINDB), invited Lew to join the National Institutes of Health (NIH) in 1962. Lew was one of perhaps 20 neuropathologists in the country at the time. Later NINDB became the National Institute of Neurological Disorders and Stroke (NINDS). NINDB was looking for a neuropathologist for the Perinatal Research Branch (PRB) headed by Dr. Heinz Berendes, M.D. When he first came to NIH, Lew was determined to build something that implemented his ideas of mapping in biological images. He had an original LINC (Laboratory INstrument Computer created at MIT with NIH funding) computer at the time. Later, Lew upgraded this to a Digital Equipment Corporation (DEC) LINC-8 . The problem: he had a microscope and he had a computer. How could he combine the two?
The first thing he wanted to be able to do was move a slide via a computer-controlled microscope stage. Initially, he was going to do it with analog feedback. He talked to Wes Clark (who had helped build the LINC computer with Charlie Molner and others). Wes convinced Lew that he really wanted a digital stage - not an analog one - so that is what Lew developed: a series of stepping-motor-controlled stages that improved with each generation. The original design connected the stage with rubber bands, which was then greatly improved with direct stepping-motor drives. Lew had also been working with Russell Kirsch and Bill Watt from the National Bureau of Standards (NBS, now the National Institute of Standards and Technology or NIST ). This early work involved describing biological images using computer picture grammars [1] that attempted to bring artificial intelligence and algorithmic methods to the description of biological images.
...
The second computer-controlled microscope project was the NCI grain counter [2] that is discussed in its own section. Advancements in electronics technology enabled us to design the grain counter using high-speed shift-register memory chips to capture X,Y coordinates from a 10 frame/second non-interlaced TV system ( Imanco Imanco Quantimet 720 ). Despite these advances, for larger image memories such as was needed for the RTPP, it would have been very difficult to implement image processing algorithms. This is because shift-register memory has delays in accessing any particular image pixel datum since the data must cycle around the circular shift register before the computer could access it. For complex algorithms with millions or billions of operations, this would be intolerable.
The culmination of these efforts was the Real Time Picture Processor (RTPP) described in journal papers [3, 4, 5, 6], as well as technical reports to be discussed and listed at the end of this history. We started this project just as the new Texas Instruments 4K bits X 1-bit dynamic RAMs (Random Access Memory - see history ofDRAM ) became available. Their availability was discovered by George Carman who proceeded to design the RTPP using these new chips. Many skilled people made this project possible: the superb computer hardware architecture work by George and the mechanical engineering work by Sprague Hazard; the coming together of the right group of people, with synergistic skills who got along as a family, at the right time when the technology and the NIH's support resources were available; the NCI's Director Seymour Perry and administrator Bill Penland gave us crucial encouragement and financial support. Dr. Perry invited us to move to NCI as the Image Processing Unit (IPU) about 1972. In projects of this type, there is a window of time when the technology is appropriate for the job. Without the 4K dynamic RAMs, the RTPP would not have been possible. We were doing cutting-edge research, but a year or two later, charge-coupled devices would make their appearance and eventually make much of our design obsolete. But that is the nature of progress.
...
One of the unique aspects of the RTPP was to implement the design as special-purpose parallel hardware with a flexible bus-architecture and a microcoded instruction set that reflected the types of operations routinely performed in image processing [3-4, TR-2, TR-7, TR-7a, TR-22]. Although other image processing computers were available, such as the ILLIAC-III , using a microcode architecture enabled an image processor to be constructed and built less expensively but with greater flexibility than building it entirely with discrete hardware. The special-purpose hardware could make real-time results possible (defined as reasonably fast enough to incorporate human feedback in tuning algorithms, such as interactively adjusting detection thresholds, etc.). A National Technical Information Service (NTIS)technical report [TR-7] describing the RTPP was one of the frequently requested reports one month as reported in their monthly newsletter for November 1976 under computer topics.
...
Scientists used the RTPP as finally constructed to analyze data in a variety of biomedical domains including optical microscope images of optical serial sections of brain tissue, stained bone marrow smears, and tissue cultures using phase contrast and differential interference optics. The latter was used in tracking cell membrane extents of macrophages in tissue culture over time as the cells tried to phagocytize various types of asbestos fibers. The goal was to better understand fiber carcinogenicity and the dynamics of fiber ingestion [8, 9, 10]. The bone marrow smear image analysis was part of my Ph.D. dissertation [11, 12, TR-653, TR-655]. The RTPP was also used for 2D electrophoretic gel images for a variety of biological materials [13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32], and for RNA electron micrographs of secondary structure, which was part of Bruce Shapiro's Ph.D. dissertation [33, 34, 35, 36, 37, 38, TR-BAS78].
Figure 1. Dr. Lewis Lipkin headed the project. His group started working on computer-controlled optical microscopy in the Perinatal Research Branch (PRB) of NINDB. The group later changed its name and institutes to the Image Processing Unit (IPU) in NCI in the Laboratory of Pathology. IPU later became the Image Processing Section (IPS) in NCI. The Section later became part of the Laboratory of Mathematical Biology (LMMB) in NCI under Dr. Charles DeLisi, Ph.D., and still later under Dr. Jacob Maizel, Ph.D.. The laboratory changed its name to the Laboratory of Experimental and Computational Biology (LECB) under Jake Maizel. The laboratory is currently refocused on nanobiology and is now called the Center for Computer Research Nanobiology Program (CCRNP) directed by Dr. Robert Blumenthal, Ph.D.(CCRNP has an additional research Web site).
...
- Lewis Lipkin, M.D., (mathematics and physical chemistry, and a neuropathologist), Head of the Image Processing Section (IPS); previously the (PRB, NINDB) and then the Image Processing Unit (IPU) in the NCI.
- Peter Lemkin, Ph.D. & M.S. EE, computer scientist and electrical engineer, IPS/NCI, and previously in (PRB, NINDB) and in IPU/NCI
- George Carman, M.S. EE, electrical engineer and computer hardware architecture, Technical Development Section (TDS), NINDB; Carman Engineering (now Lucidyne Corp ).
- Morton Schultz, B.S. EE, electrical engineer, IPS/NCI, and previously in IPU/NCI
- Bruce Shapiro, Ph.D., B.S. math & physics, computer scientist, IPS/NCI, and previously in (PRB, NINDB),and in IPU/NCI
- Sprague Hazard, mechanical engineer (contractor consultant)
- Peter Kaiser, B.S. CS, computer scientist (IPU) in the NCI
- Earl Smith, M.S. CS, computer scientist (IPU) in the NCI
- Dan Kilgore, B.S. EE, computer programmer [Digital Equipment Corp Corp (DEC) software engineer]
- Tom Duval and later Jim Camper, electronics technicians - helped construct the RTPP racks, and power-supplies cabinets
- Cambion Corporation, wire-wrapped the remaining 63 buffer memory boards and the back-planes
...
- Lewis Lipkin: optical microscopy of serial brain sections and macrophage motility measurements with asbestos
- Peter Lemkin: bone marrow smear analysis, 2D gel electrophoresis
- Bruce Shapiro: RNA secondary structure of electron micrograph
- Carl Merril: NIMH/NIH - 2-dimensional (2D) gel electrophoresis, E.coli mutants and macrophages with asbestos
- Jacob Maizel: NICHD/NIH, with Bruce Shapiro - RNA electron microscopy of secondary structure
- Eric Lester: NCI, U. Chicago, and oncology practice - 2D gel electrophoresis on human leukemia
- Steve Aley and Russell Howard: NIAID/NIH - 2D gel electrophoresis of Plasmodium knowlesi clones
- Peter Wirth and Snorri Thorgeirsson: NCI/NIH - 2D gel electrophoresis on liver cell lines
- Peter Sonderegger: NICHD/NIH and U. Zurich - 2D gel electrophoresis of axonal proteins of sensory and motor neurons
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
...
The design of the RTPP was presented at the 1973 Asilomar Third Engineering Foundation Conference on Automated Cytology and published in 1974 [3-4]. This conference and a subsequent automated cytology workshop concentrated on the two solutions then available: image processing and pattern recognition of cell images, and the evolving field of flow cytometry. NIH was funding both fields. During this time we developed plans for integrating artificial intelligence techniques for understanding and analyzing biological materials and systems incorporating the RTPP, and these were also presented at the Asilomar workshop [5, TR-15].
...
Not only did Cambion build the buffer memory boards, but using their standard technology, they also wired much of the backplanes. Their technology was an integrated system, and had been developed for rapid prototype construction in aerospace projects. It included cards, sockets, and racks. The system would not have worked had the parts been obtained from different vendors. By adapting Cambion's standards, we were able to take advantage of the reliability Cambion had developed for this kind of work. After Cambion created the boards and backplanes, our electronics technicians assembled them into several cabinets of 19" vertical racks including one cabinet for the power supplies. The buffer memories were 16 cards to a rack, with four racks. To avoid overheating, the cards were inserted in every other slot. Then the equipment was shipped to George in Oregon to finish construction and debugging. We had purchased a PDP8e for him to use in developing, debugging, and testing the interface. The computer was also critical for George to create various software tools to help manage the project. These included a wirewrap database program that could take pairs of (drawing #, chip #, pin #) triples that indicated a pair of wires to be connected using a technique called "wirewrap." This methodology was critical since a single buffer memory card was described in a large number of blueprints and it would be difficult to keep straight which pins connected to other pins in this complex global diagram. George then wrote additional software to translate these pairs to the standard lists that Cambion required. In a biomedical image processing and electronics conference, George's triple notation and his new way to handle the increasing complexity of multiple drawing wiring lists received a good reception from some of the developers of VHDL (a hardware description language). Because of space limitations, George put the PDP8e into a closet of his house with additional AC cooling. The PDP8e at that time cost more than his house. Today, the most inexpensive computers are many orders of magnitude more powerful than the PDP8e at a small fraction of their cost.
...
The Axiomat was a dream microscope. (A photograph is available on Zeiss's microscope history Web page.) The microscope complex grew in the sense that as we wanted more and more control of the microscope functionality, we added it. In addition to control of the stage and control of the Z-axis, we also wanted control of the frequency of light that went through it. Although we experimented with various color selection methods, we settled for using interference filters. The RTPP and the microscope were controlled in real-time by a polling routine in BMON2 with the (X,Y,Z) direction control switches, A/Ds, and other states available for programs needing this data. Of course Lew Lipkin's pick-list idea was implemented and was part of BMON2.
...
The software control program for the buffer memory I constructed on the PDP8e was called BMON2 (the Buffer Memory Monitor System) [40, TR-21, TR-21b, TR-23] and written in Fortran-II. BMON2, in addition to interfacing with the RTPP, also allowed running other programs to be batched to analyze the data. Given that the PDP8e had 32K words of memory, this was critical for doing complex sequential operations and for easily writing new RTPP applications. A Fortran-II library that could interface with the RTPP, BMOMNI [TR-23], allowed these other programs to access the RTPP as required. (See discussion on Fortran-II in the section on the grain counter. This shows the BMOMNI Fortran code.) BMON2 could capture and display images and do many image processing operations on the PDP8e. Another program called FLICKER [13] ran on the PDP8e and was used to analyze 2D gel images visually by alternately displaying one movable image on the video screen relative to another that was held in a constant screen position. Later, it allowed the comparison of two saved images as well. So a set of images could be compared against a reference sample. Some of the ideas on using flickering images to detect subtle differences in image matching were suggested by Bernice Lipkin, who is an expert in psychopictorics [41]. A third-generation version of FLICKER is available as open-source software at http://open2dprot.sourceforge.net/Flicker .
| Anchor | ||||
|---|---|---|---|---|
|
...
A Digital Equipment Corporation DECsystem-2020 was installed in the laboratory after we moved to the Park Building. We had been using the NIH's Division of Computer Research and Technology (DCRT) [now the Center for Information Technology (CIT)] DECsystem-10 time-shared system. As we used TOPS-10 operating system on the DECsystem-10, we installed TOPS-10 on the new DECsystem-2020. Bruce Shapiro had implemented a message-switching high-speed 9600-baud (normal speed was 300 or 1200 baud at the time) serial line multiplexor so we could move images and data to/from the DCRT system. However, the costs for the increasing amount of time we used on the DCRT system was escalating. For a cost comparable to renting time over a few years, we could purchase a dedicated system and have more compute power as well. So NCI supported us in purchasing the DECsystem-2020. This was a DEC Unibus system, which meant we could interface our hardware to this then-powerful 36-bit computer. In hindsight, this was one of the best procurements that Lew made. It offered us vastly better opportunities to interact with and manage the data that would not have been possible with a 9600 baud serial line. We could write software in the SAIL language, which meant we would have much more expressive power than we had with the PDP8e or PDP11 computers and could apply more advanced algorithms. This made a real difference in the productivity in analyzing real data with powerful algorithms.
...
In addition to the RTPP, we acquired Comtal image processor systems that had Q-bus type PDP11 interfaces. These in turn were interfaced to a PDP11/40 computer that was connected to the DECsystem-2020 via software called SPIDER, a virtual device driver network. SPIDER allowed PDP11 computers to be accessed from the DECsystem2020 without writing a new DECsystem-2020 driver for each new PDP11 device. Bruce Shapiro, our expert on PDP11s, wrote a time-shared packet switcher on the PDP11/40 to connect PDP11 devices to this network. I wrote the device driver on the DECsystem-2020 to access these devices and make them available for DECsystem-2020 application software. Images acquired using the RTPP could be analyzed on the Comtals; Bruce used this to help analyze his nucleic acid electron micrographs.
...
Many software analysis systems were developed using the RTPP, especially in the area of 2D gels with the GELLAB-I system [13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,24, 25, 26, 27, 28, 29, 30, 31, 32], a 2D gel exploratory data analysis system integrating the image-processing with statistical databases for multiple samples (myself); and RNA electron micrographs of secondary structure [10, 11, 32, 39, 48, 49] (Bruce Shapiro). After the RTPP was decommissioned, GELLAB-I was redeveloped as a portable software system using Unix/C/X-windows and was called GELLAB-II [42, 43, 44, 45] (see Lemkin's History of GELLAB for more details, references, and history of GELLAB-II). Much of the work with GELLAB-I and GELLAB-II in exploratory data analysis led to its application to the DNA microarray domain (see http://maexplorer.sourceforge.net/ ) MAExplorer []. A third-generation instantiation of this data-mining system is part of the Open2Dprot open-source project at at http://open2dprot.sourceforge.net/ with the goal of extending proteomics data mining to 2D LC-MS, protein-arrays. Bruce went on to develop other RNA analysis software [35, 39, 47, 48, 49, 50], leading to the StructureLab project [50] and related RNA structure analysis (see his RNA structure research group ).
6.
| Anchor | ||||
|---|---|---|---|---|
|
...
| Anchor | ||||
|---|---|---|---|---|
|
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
Anchor RTPP-Quantimet-PDP8e-blockDiagram RTPP-Quantimet-PDP8e-blockDiagram
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
...
Anchor ControlDesk ControlDesk
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
...
Anchor PhotoRTPPandConsole PhotoRTPPandConsole
| Anchor | ||||
|---|---|---|---|---|
|
The buffer memory image cards
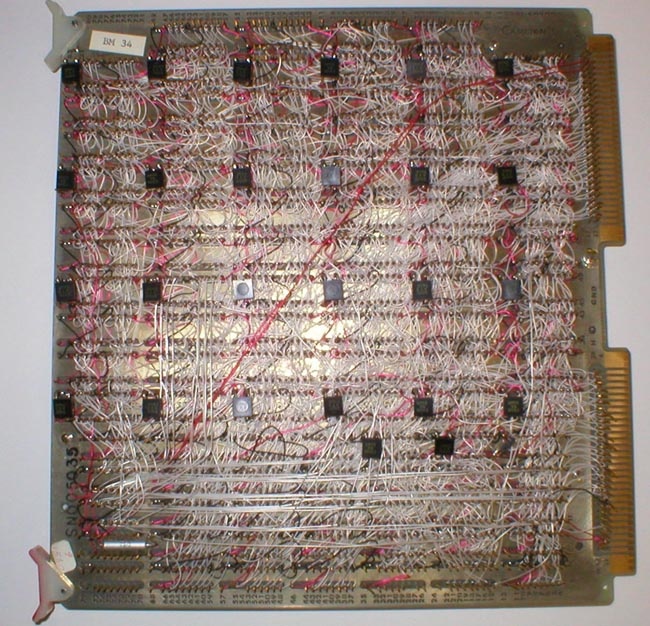
The sample card shown below (front and back) illustrates the complexity of building large memory cards back in the early 1970s (Figures 8 through 10). Increases in chip memory capacity enabled enough memory to be put on a few cards to represent images with detail sufficient for meaningful biological image analysis. Before these new memory chips (the Texas Instruments TMS 4030 chip was a 4Kx1 bit dynamic RAM) were available, we had planned to construct the buffer memories using static shift-register memory chips (as George had done in the grain counter system). Total memory pixel size was 1 megabyte for the 64 boards - today 1 gigabyte of memory the size of a thumbnail sells for under $25 (on sale). Not only would the static shift-register memory chips have been more expensive, but also they also would not have had the random access performance we required in image processing, creating an inadequate design. The shift registers would have led to a slower, much smaller memory using more power for the same cost. The image pixels size and number of images in the buffer memories would have been less and many of the powerful software applications we used the RTPP for would not have worked as well or even been developed.
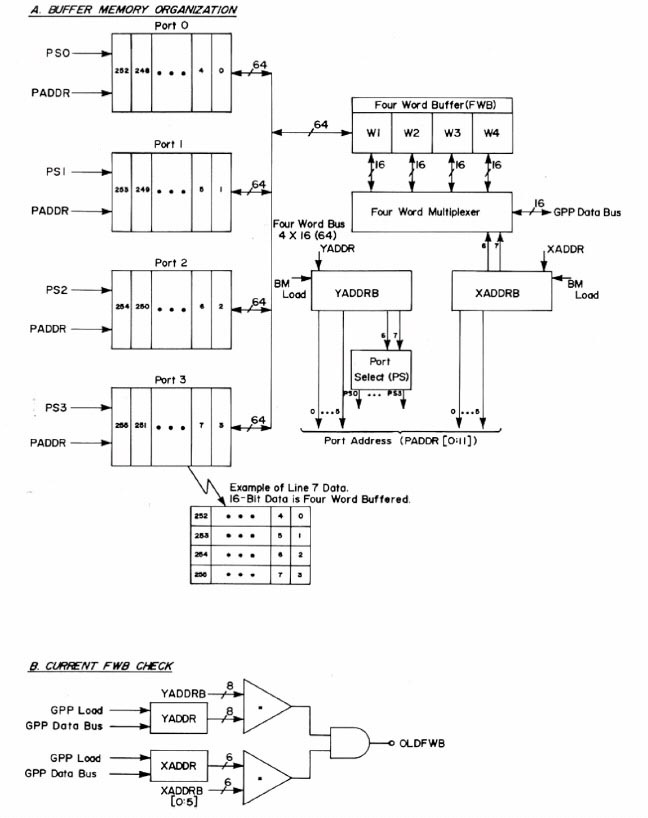
During this time, we had the conviction, led by Lew Lipkin and George Carman, that anything that we wanted to be do in software could be done by a series of sequential gates. These could be proved Boolean algebraically correct using Karnaugh Maps, hardware finite state machines, and related techniques. George had just taken a microprogramming design course as part of his masters degree in computer hardware architecture and the design of buffer memories and the General Picture Processor (GPP) were perfect test beds in which to try out these new design principles which were relatively new for projects like this. Some of the design diagrams are shown in Figures 11 through 14 (from the Carman [4] paper). Figure 15 shows some examples of GPP microprogrammed instructions for manipulating the buffer memory data. The design was further described in some of the technical reports [TR-7, TR-7a, TR-16, TR-21, TR-21b, TR-22] listed at the end of this history. Because we were prototyping the system, the card was constructed using wire wrapping rather than multilayer printed circuit boards. A commercial version would have used printed circuit boards, but would only have been economically feasible if many copies of the RTPP were produced. Using complex multi-level printed circuit boards is generally too expensive for a research lab.
| Anchor | ||||
|---|---|---|---|---|
|
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
...
Anchor TMS4030chip TMS4030chip
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
Anchor Fig-RTPP-BufferMemory-control-logic Fig-RTPP-BufferMemory-control-logic
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
| Anchor | ||||
|---|---|---|---|---|
|
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
| Div | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||
|
| Anchor | ||||
|---|---|---|---|---|
|
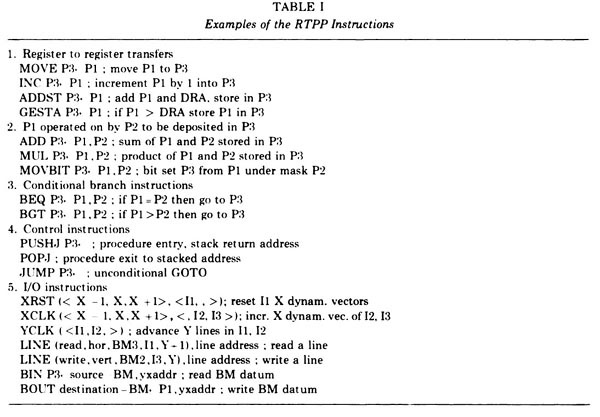
Figure 15. The Examples of RTPP instructions for the GPP (reproduced with permission from J. Histochem. Cytochem. [4], 1974). The Pi refers to a 3x3 pixel neighborhood that would be tessellated through the entire image. The GPP instructions [A HREF="#TR-22">TR-22] could be compiled by the GPPASM [A HREF="#TR-16">TR-16] assembler program running on the PDP8e and then loaded into the GPP instruction memory. A debugger for the GPP was DDTG that ran on the PDP8e [A HREF="#TR-2">TR-2] , but controlled the GPP and buffer memories. We had also been evaluating collaborating on the construction of a a MAINSAIL(R) compiler to generate GPP assembly code so we could program the RTPP in a SAIL-like language.Anchor Fig-RTPP-Examples-of-GPASM-code Fig-RTPP-Examples-of-GPASM-code
...
Another of the projects Lew and I were also working on was the construction of a 3D brain atlas of an anolus lizard brain using aligned microtomed serial-section images. (Some of the goals for that project were similar to that of the National Library of Medicine's Visible Human. However, this was well before adequate computer technology and resources were available to implement such an atlas.) We wrote a PDP8e program to move the optical microscope stage in (X,Y) while keeping the center of the brain in the scanned visual center of the image field as we switched slides in a series of microtomed serial sections (the Z-axis control was not used in this operation). Each centered image could then be captured and saved as a disk file. The next section visible on the TV camera was compared with the image previously captured and stored in the image buffer memory. This procedure was iteratively repeated with subsequent slides to capture a sequence of aligned serial sections from a set of slides. The buffer memory data was saved at each point on 9-track magnetic tape. (This alignment software subsequently led to our involvement with 2D electrophoretic gels analysis [discussed below].) The set of serial sections could then be re-accessed sequentially to step through the brain slices centered at the selected point. We could generate movie loops with the BMON2 software to move virtually up and down through the serial sections at various frame rates. Because we could store sixteen 256x256 images in the buffer memory, the loops could have up to 16 sections and allowed us to visualize the 3D structure of part of the brain.
| Anchor | ||||
|---|---|---|---|---|
|
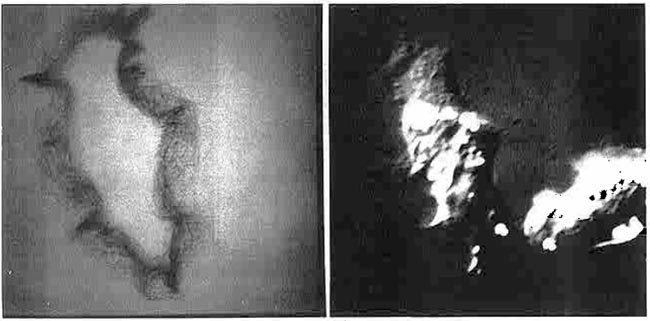
Figure 17. (Left) Boundary trace transform (BTT) of 233 images of 15-second interval scans of a single living P388D1 macrophage-like cell (left) (reproduced with permission from Environmental Health Perspectives, 1980 [9]). The boundaries for this set of images were traced by hand using a graphics tablet connected to the RTPP. The BTT is a 2D boundary frequency histogram where darker (higher frequency) pixels indicate boundaries of the cell that are more adherent to the glass slide and have less motility. BMON2 captured the set of images data and double buffered them to 9-track magnetic tape, with a 15-second image-sampling interval between scans. (Right) Illustration of a single cell captured by the RTPP using differential interference optics. The algorithm is described in several papers [8, 9]. Movies were made on the RTPP of a subsequence of 16 of the images in which you could see the cell trying to ingest the fiber, the fiber breaking through the other side of the cell (membrane), and then the cell backing up to try to reingest that part of the fiber, etc. These images allowed us to create a probability distribution of adhesion strength for boundary points of amoeboid cells. Points with low probability would flutter and extend. Looking at individual boundaries would not reveal this type of information; it was the ability to integrate large quantities of data that allowed these patterns to be detected. This was the way we thought the system should have been used - finding interesting results when no other method for doing so was possible.
...
The FLICKER software allowed users to measure spots as a set of (X,Y) image positions and integrated-densities for each sample. We manually recorded this data for later analysis, manually defining spots to be corresponding if they aligned well with FLICKER.
Figure 18. This was the alternative image acquisition setup. (Reproduced from a figure with permission from a reprint from Environmental Health Perspectives, 1980 [9].) A high-quality uniform illumination Aristo light box and Quantimet Vidicon TV camera were used for non-microscope images. A gel autoradiograph, wet 2D gel, electron micrograph, or other transparent sample object was placed on the light box and scanned using the RTPP/Vidicon camera. A NBS standard neutral density step wedge was placed on the bottom of the scan area. This was captured along with the image and was used to calibrate the grayscale pixel data to optical density using a piecewise linear curve fitting algorithm (see [9] for an example). As an aside, the electronics workbench area in the background was where many of the local RTPP electronics assembly was performed in the Park Building.Anchor lightBoxCameraStation lightBoxCameraStation
...
Eric painstakingly generated paired data for about 1,400 spots from the segmented images and their paired spot lists for a few gels. Because of the difficulty in matching or pairing spots between gels, I also developed spot-pairing and spot "landmarking" programs, which ran interactively on the RTPP/PDP8e. (Landmarking is visually identifying a set of spots that are common in both the reference gel and each additional gel sample so spot pairing could proceed.) Eric then subjected the data to SPSS statistical analysis with encouraging results (t-Tests, Spearman correlation, ANOVA) and it was published in [14] and enhancements in [15,18-21]. All three of these programs were rewritten in SAIL for the DECsystem-2020. At this point we realized that we wanted to build a database containing large numbers of gels to detect marker proteins or classify samples by protein pattern signatures indicating states of differentiation, disease conditions, or other experimental conditions. This was the basis of GELLAB-I.
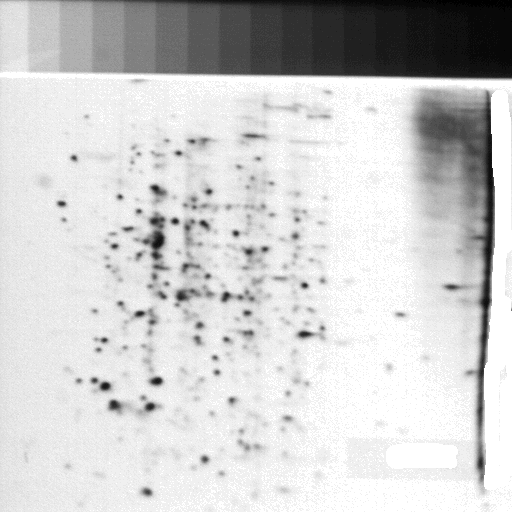
This led to the SAIL program CGELP, in GELLAB-I, to construct a composite gel database with a virtual reference gel [14-18,20,30] and the addition of many more statistical methods. (The subsequent Unix version was called CGELP2 [30, 42, 43, 44, 45] where additional statistical exploratory data analysis methods were added - see [history of GELLAB ].) Spots of a set of N-1 gels would be matched to one of the gels called a reference gel and spots missing in the physical reference gel would be extrapolated into the reference gel. Figure 19 shows the reference gel 324.1 that was an acute myeloid leukemia (AML) gel, scanned with the RTPP. This reference gel was used in many of the leukemia databases to tie the data together [14, 15, 16, 17, 18, 22]. The collection of SAIL programs, as well as their RTPP interface, was called GELLAB-I. After leaving NIH (for the University of Chicago), Eric would fly back to work in our laboratory to do marathon late-night landmarking sessions to help generate these databases of large numbers of 2D leukemia gels. Over the years, we had built various databases with over 400 gel samples. The leukemia database had over 130 samples. Some of these gel sample images are available on the bioinformatics.org/lecb2dgeldb open source repository. This early research led to my interest in exploratory data analysis and future work with microarrays with MAExplorer.sourceforge.net [46], and proteomics exploratory data analysis using open2dprot.sourceforge.net .
Peter Sonderegger, while a post-doc at the National Institute of Child Health and Development (NICHD), used the GELLAB-I system with the RTPP to investigate how the expression of axonal proteins of sensory and motor neurons was influenced by non-neuronal cells [27, 29, 45]. At the same time we were investigating the feasibility of porting GELLAB-I (written in SAIL) to the PASCAL computer language. The inflexibility of PASCAL eventually led us to convert GELLAB-I to the portable C/UNIX/X-windows environment called GELLAB-II [42, 43, 44, 45] The DECsystem-10 SAIL version, GELLAB-I, was exported to research labs at Univ. of Chicago (Eric Lester) and Univ. of Kiel (Heinz Busse). The Unix version, GELLAB-II, was exported to a number of research labs around the world (CDC with Jim Myrick [44], Univ. Zurich with Peter Sonderegger, Agr. Univ. Norway with Trygve Krekling, and others) and led to a commercial subset version for Windows PCs called GELLAB-II++ by CSPI/Scanalytics.
| Anchor | ||||
|---|---|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|
...
Partly as a result of this collaboration, Jake became more interested in computing, which led him to purchase one of the early DEC VAX computers for his lab. As Jake became more involved with computational techniques, he realized the general importance, as did Lew, of using computers in biology. Eventually, this interest culminated with the National Cancer Institute's purchase of a Cray XMP supercomputer, and the establishment of what is today the Advanced Biomedical Computer Center in Frederick. The Cray XMP, at that time, was the first supercomputer in the world solely dedicated to biomedical research. Thus, the impact of the RTPP continues to be felt in the biological computation sciences 30 years later.
| Anchor | ||||
|---|---|---|---|---|
|
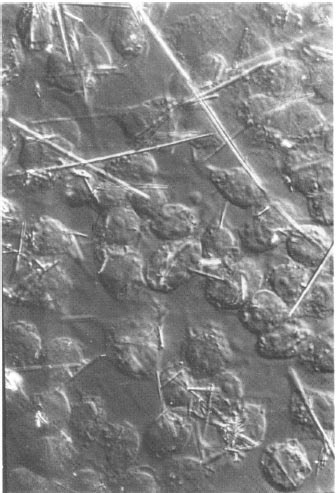
Figure 20. One of the early RNA electron micrographs scanned with the vidicon/RTPP system (Jacob Maizel, Bruce Shapiro, and Lewis Lipkin) [33, 34, 39, 48, 49]. The sample was adenovirus type 2 messenger RNA. Bruce developed boundary segmenters and boundary shape descriptors that could map electron micrograph data to the secondary structure.
...
| Anchor | ||||
|---|---|---|---|---|
|
Lipkin, L.E., Watt, W.C., Kirsch, R.A.: The analysis, synthesis, and description of biological images. Ann N Y Acad Sci. 128(3): 984-1012, 1966.Anchor Lipkin66 Lipkin66
Lipkin, L.E., Lemkin, P.F., Carman, G.: Automated autoradiographic grain counting in human determined context. J. Histochem. Cytochem. 22(7): 755-765, 1974. (PDF )Anchor Lipkin74 Lipkin74
Lemkin, P.F., Carman, G., Lipkin, L., Shapiro, B., Schultz, M., Kaiser, P.: A real time picture processor for use in biologic cell identification. I. System design. J. Histochem. Cytochem. 22(7): 725-731, 1974. (PDF )Anchor RTPP-JHC-I-paper RTPP-JHC-I-paper
Carman, G., Lemkin, P.F., Lipkin, L., Shapiro, B., Schultz, M., Kaiser, P.: A real time picture processor for use in biologic cell identification. II. Hardware implementation. J. Histochem. Cytochem. 22(7): 732-740, 1974. (PDF )Anchor RTPP-JHC-II-paper RTPP-JHC-II-paper
Shapiro, B., Lemkin, P.F., Lipkin, L.: The application of artificial intelligence techniques to biologic cell identification. J. Histochem. Cytochem. 22(7): 741-750, 1974. (PDF )Anchor PRDL-ECR-paper PRDL-ECR-paper
Schultz, M.L., Lipkin, L.E., Wade, M.J., Lemkin, P.F., Carman, G.M.: High resolution shading correction. J. Histochem. Cytochem.22(7): 751-754, 1974. (PDF )Anchor Schultz74 Schultz74
Shapiro, H.M., Bryan, S.D., Lipkin, L.E., Stein, P.G., Lemkin, P.F.: Computer-aided microspectrophotometry of biological specimens. Exp Cell Res. 67(1): 81-89, 1971.Anchor Shapiro71 Shapiro71
Lemkin, P.F.: The boundary trace transform: An edge and region enhancement transform. Comp. Graphics Image Processing 9: 150-165, 1979.Anchor Lemkin79a Lemkin79a
Lemkin, P.F., Lipkin, L., Merril, C., Shiffrin, S.: Protein abnormalities in macrophages bearing asbestos. Environ. Health Perspect. 34: 5-89, 1980. (PDF)Anchor Lemkin80a Lemkin80a
Lipkin, L.E.: Cellular effects of asbestos and other fibers: correlations with in vivo induction of pleural sarcoma. Environ. Health Perspect. 34:91-102, 1980. (PDF)Anchor Lemkin80b Lemkin80b
Lemkin, P.F.: An approach to region splitting. Comp. Graphics Image Processing 10: 281-288, 1979.Anchor Lemkin79b Lemkin79b
Lemkin, P.F., Lipkin, L.: Use of the positive difference transform for RBC elimination in bone marrow smear images. Anal. Quant. Cytol. 1(1): 67-76, 1979.Anchor Lemkin79c Lemkin79c
Lemkin, P.F., Merril, C., Lipkin, L., Van Keuren, M., Oertel, W., Shapiro, B., Wade, M., Schultz, M., Smith, E.: Software aids for the analysis of 2D gel electrophoresis images. Comput. Biomed. Res. 12: 517-544, 1979.Anchor Lemkin79d Lemkin79d
Lester, E.P., Lemkin, P.F., Cooper, H.L., Lipkin, L.E.: Computer-assisted analysis of two-dimensional electrophoresis of human peripheral blood lymphocytes. Clin. Chem. 26: 1392-1402, 1980. (PDF )Anchor Lemkin80c Lemkin80c
Lipkin, L.E., Lemkin, P.F.: Data base techniques for multiple PAGE (2D gel) analysis. Clin. Chem. 26: 1403-1413, 1980. (PDF )Anchor Lemkin80d Lemkin80d
Lemkin, P.F., Lipkin, L.: GELLAB: A computer system for 2D gel electrophoresis analysis. I. Segmentation and preliminaries. Comput. Biomed. Res. 14: 272-297, 1981.Anchor Lemkin81a Lemkin81a
Lemkin, P.F., Lipkin, L.: GELLAB: A computer system for 2D gel electrophoresis analysis. II. Spot pairing. Comput. Biomed. Res. 14: 355-380, 1981.Anchor Lemkin81b Lemkin81b
Lemkin, P.F., Lipkin, L.: GELLAB: A computer system for 2D gel electrophoresis analysis. III. Multiple gel analysis. Comput. Biomed. Res. 14: 407-446, 1981.Anchor Lemkin81c Lemkin81c
Lester, E.P., Lemkin, P.F., Lipkin, L.E.: New dimensions in protein analysis - 2D gels coming of age through Image Processing. Anal. Chem. 53: 390A-397A, 1981.Anchor Lemkin81d Lemkin81d
Lester, E.P., Lemkin, P.F., Lipkin, L.E., Cooper, H.L.: A two-dimensional electrophoretic analysis of protein synthesis in resting and growing lymphocytes in vitro. J. Immunol. 126: 1428-1434, 1981.Anchor Lemkin81e Lemkin81e
Lemkin, P.F., Lipkin, L.E., Lester, E.P.: Some extensions to the GELLAB 2D electrophoresis gel analysis system. Clin. Chem. 28: 840-849, 1982. (PDF )Anchor Lemkin82a Lemkin82a
Lester, E.P., Lemkin, P.F., Lipkin, L.E.: A two-dimensional gel analysis of autologous T and B lymphoblastoid cell lines. Clin. Chem.28: 828-839, 1982. (PDF )Anchor Lester82a Lester82a
Lester, E.P., Lemkin, P.F., Lowery, J.F., Lipkin, L.E.: Human leukemias: A preliminary 2D electrophoretic analysis. Electrophoresis 3: 364-375, 1982.Anchor Lester82b Lester82b
Howard, R.J., Aley, S.B., Lemkin, P.F.: High resolution comparison of Plasmodium Knowlesi clones of different variant antigen phenotypes by 2D gel electrophoresis and computer analysis. Electrophoresis 4: 420-427, 1983.Anchor Howard83 Howard83
Lemkin, P.F., Lipkin, L.E.: 2D Electrophoresis gel data base analysis: Aspects of data structures and search strategies in GELLAB. Electrophoresis 4: 71-81, 1983.Anchor Lemkin83a Lemkin83a
Lester, E.P., Lemkin, P.F., Lipkin, L.E.: States of differentiation in leukemias: A 2D gel analysis. In Rowley, J. D. and Ultmann, J. E. (Eds.): Proceedings of 5th Annual Bristol Myers Symposium on Cancer Research. Chromosomes and Cancer: From Molecules to Man. New York, Academic Press, 1983, pp. 226-245.Anchor Lester83a Lester83a
Lemkin, P.F., Sonderegger, P., Lipkin, L.: Identification of coordinate pairs of polypeptides: A technique for screening of putative precursor product pairs in 2D gels.Clin. Chem. 30: 1965-1971, 1984. (PDF )Anchor Lemkin84a Lemkin84a
Lester, E.P., Lemkin, P F., Lipkin, L.E.: Protein indexing in leukemias and lymphomas. Ann. N.Y. Acad. Sci. 428: 158-172, 1984.Anchor Lester84a Lester84a
Sonderegger, P., Lemkin, P.F., Lipkin, L., Nelson, P.: Differential modulation of the expression of axonal proteins by non-neuronal cells and the peripheral and central nervous system. EMBO J. 4: 1395-1401, 1985. (PDF)Anchor Sonderegger85 Sonderegger85
Lemkin, P.F., Lipkin, L.E.: GELLAB: Multiple 2D electrophoretic gel analysis. In Allen, R. and Arnaud (Eds.): Electrophoresis '81. New York, W. De Gruyter, 1981, pp. 401-411.Anchor Lemkin81f Lemkin81f
Lemkin, P.F. , Lipkin, L.E.: Database techniques for 2D electrophoretic gel analysis. In Geisow, M. and Barrett, A. (Eds.): Computing in Biological Science. North Holland, Elsevier, 1983, pp. 181-226.Anchor Lemkin83b Lemkin83b
Lester, E.P., Lemkin, P.F.: A 'GELLAB' computer assisted 2D gel analysis of states of differentiation in hematopoietic cells. In Neuhoff, V. (Ed.): Electrophoresis '84. Chemie, Springer-Verlag, 1984, pp. 309-311.Anchor Lester84a Lester84a
Lemkin, P.F., Shapiro, B., Lipkin, L., Maizel, J., Sklansky, J., Schultz, M.: Preprocessing of electron micrographs of nucleic acid molecules for automatic analysis by computer. II. Noise removal and gap filling. Comput. Biomed. Res. 12: 615-630, 1979.Anchor Lemkin79d Lemkin79d
Lipkin, L., Lemkin, P.F., Shapiro, B., Sklansky, J.: Preprocessing of electron micrographs of nucleic acid molecules for automatic analysis by computer. Comput. Biomed. Res. 12: 279-289, 1979.Anchor Lipkin79b Lipkin79b
Shapiro, B., Lipkin L.: The circle transform, an articulable shape descriptor. Comput. Biomed. Res. 10: 511-28, 1977.Anchor Shapiro77a Shapiro77a
Shapiro, B.: Language processor generation with BNF inputs: methods and implementation. Comp. Programs. Biomedicine 7:85-98, 1977.Anchor Shapiro77b Shapiro77b
Shapiro, B., Pisa, J., Sklansky, J.: Skeletons from sequential boundary data. Proc. Intl. Conf. On Pattern Recognition and Image Processing. IEEE Comp. Soc. Press, Los Angeles, CA., 265-270, 1979.Anchor Shapiro79a Shapiro79a
Shapiro, B., Pisa, J., Sklansky, J.: Skeleton generation from xy boundary sequences. Comp. Vision Graphics Image Processing 15(2) 136-153, 1981.Anchor Shapiro81a Shapiro81a
Shapiro, B.S., Lipkin, L.E., Maizel, J.V.: Computerized generation of secondary structure maps for nucleic acids. Comp. Biomed. Res.12(6):545-568, 1979.Anchor Shapiro79b Shapiro79b
Lemkin, P.F., Lipkin, L.: BMON2 - A distributed monitor system for biological image processing. Computer Programs in Biomedicine 11: 21-42, 1980. (PDF)Reprinted from COMPUTER METHODS AND PROGRAMS IN BIOMEDICINE, Vol 11, Lemkin PD and LipkinL, BMON2 - A distributed monitor system for biological image processing, Pages 21-42, Copyright (1980), with permission from Elsevier.Anchor BMON2-CPIB-paper BMON2-CPIB-paper
Lipkin, B.S., Rosenfeld, A. (Eds): Picture Processing and Psychopictorics. Academic Press, New York, 1970, pps 526.Anchor Lipkin70 Lipkin70
Lemkin, P.F.: GELLAB-II: A workstation based 2D electrophoresis gel analysis system. In Endler, T. and Hanash, S. (Eds.): Proceedings of 2D Electrophoresis. West Germany, VCH Press, 1989, pp. 52-57. (This was the announcement of GELLAB-II)Anchor Lipkin89g Lipkin89g
Lemkin, P.F., Lester, E.P.: Database and search techniques for 2D gel protein data: A comparison of paradigms for exploratory data analysis and prospects for biological modeling. Electrophoresis 10(2): 122-140, 1989.Anchor Lipkin89h Lipkin89h
Robinson, M.K., Myrick, J.E., Henderson, L.O., Coles, C.D., Powell, M.K., Orr, G.A., Lemkin, P.F.: Two-dimensional protein electrophoresis and multiple hypothesis testing to detect potential serum protein biomarkers in children with fetal alcohol syndrome. Electrophoresis 16: 1176-1183, 1995.Anchor Robinson95 Robinson95
Stoeckli, E.T., Lemkin, P.F., Kuhn, T.B., Ruegg, M.A., Heller, M., Sonderegger, P.: Identification of proteins secreted from axons of embryonic dorsal-root-ganglia neurons. Eur. J. Biochem. 180: 249-258, 1989.Anchor Stoeckli89 Stoeckli89
Lemkin, P.F., Thornwall, G., Walton, K., Hennighausen, L: The Microarray Explorer tool for data mining of cDNA microarrays - application for the mammary gland, Nucleic Acids Res. 20(22): 4452-4459, 2000.Anchor Lemkin00a Lemkin00a
Shapiro, B.A.: An algorithm for comparing multiple RNA secondary structures. Comput. Appl. Biosci. 4(3): 387-393, 1988.Anchor Shapiro88 Shapiro88
Margalit, H., Shapiro, B.A., Oppenheim, A.B., Maizel, J.V. Jr.: Detection of common motifs in RNA secondary structures. Nucleic Acids Res. 17(12): 4829-4845, 1989.Anchor Margalit89 Margalit89
Le, S.Y., Owens, J., Nussinov, R., Chen, J.H., Shapiro, B., Maizel, J.V.: RNA secondary structures: comparison and determination of frequently recurring substructures by consensus. Comput. Appl. Biosci. 5(3): 205-210, 1989.Anchor Le89 Le89
Shapiro, B.A., Kasprzak, W.: STRUCTURELAB: a heterogeneous bioinformatics system for RNA structure analysis. J Mol. Graph.14(4): 194-205, 222-224, 1996.Anchor Shapiro96 Shapiro96
...
These reports were archived in the U.S. Department of Commerce's National Technical Information Service (NTIS) in Springfield VA. They currently offer online reports from 1990 on their web site http://www.ntis.gov/, although older reports can be ordered. Some additional reports appeared as University of Maryland Computer Science Center technical reports.
TR-2. Lemkin, P.F.: DDTG - Functional specification for the RTPP monitor. NCI/IP TR-2, 2-5-1976. NTIS Accession No. PB250726/AS, Springfield, VA, 1976, 90 pp. (Also in DECUS No. 8-823). (PDF)Anchor TR-2-report TR-2-report
TR-7. Lemkin, P.F., Carman, G., Lipkin, L., Shapiro, B., Schultz, M.: Real time picture processor: Description and specification. NCI/IP TR-7, 2-31-1976. NTIS Accession No. PB252268/AS, Springfield, VA, 1976, 139 pp. (PDF)Anchor TR-7-report TR-7-report
TR-7a. Lemkin, P.F.: Real Time Picture Processor: Description and specification. TR-7a, 6-23-1977. NTIS Accession No. PB269600/AS, Springfield, VA, 1977, 185 pp. (PDF TR-7a)Anchor TR-7a-report TR-7a-report
TR-8. Lemkin, P.F., Shapiro, B., Gordon, R., Lipkin, L.: PROC10 - An image processing system for the PDP10: Description and specification. NCI/IP TR-8, 12-16-1976. NTIS Accession No. PB261535/AS. Springfield, Va., 1976, 53 pp. (Also in DECUS No. 10-270). (PDF)Anchor TR-8-report TR-8-report
TR-15. Shapiro, B., Lemkin, P.F.: PRDL - Procedural Description Language. NCI/IP TR-15, 10-10-1977. NTIS Accession No. PB273112/AS Springfield, Va., 1977, 23 pp. (PDF)Anchor TR-15-report TR-15-report
TR-16. Lemkin, P.F., Shapiro, B., Schultz, M., Lipkin, L., Carman, G.: GPPASM - A PDP8e assembler for the General Picture Processor. NCI/IP TR-16, 12-15-1976. NTIS Accession No. PB261537/AS, Springfield, VA, 1976, 45 pp. (PDF)Anchor TR-16-report TR-16-report
TR-21. Lemkin, P.F.: Buffer memory monitor system for interactive image processing. NCI/IP TR-21, 3-31-1976. NTIS Accession No. PB261536/AS, Springfield, VA, 1976, 26 pp. (PDF)Anchor TR-21-report TR-21-report
TR21-b. Lemkin, P.F.: BMON2 - buffer memory monitor system for interactive image processing. NCI/IP TR-21b, 3-17-1978. NTIS Accession No. PB269642/AS, Springfield, VA, 1978, 112 pp. (PDF)Anchor TR-21b-report TR-21b-report
TR-22. Carman, G., Lemkin, P.F., Schultz, M., Lipkin, L., Shapiro, B.: Microprogram control architecture for the General Picture Processor. NCI/IP TR-22, 4-22-1977. NTIS Accession No. PB269762/AS, Springfield, VA, 1977, 35 pp. (PDF)Anchor TR-22-report TR-22-report
TR-23. Lemkin, P.F.: BMOMNI - Fortran interface program for the RTPP buffer memory, Quantimet and control desk. NCI/IP TR-23, 12-14-1976. NTIS Accession No. PB261538/AS, Springfield, VA, 1976, 10 pp. (PDF)Anchor TR-23-report TR-23-report
TR-472. Shapiro, B.: The use of orthogonal expansion for biological shape description. College Park, MD, University of Maryland Computer Science Center TR-472, Aug. 1976, pp 30. TR-653. Lemkin, P.F.: Bone marrow smear image analysis. College Park, MD, University of Maryland Computer Science Center TR-653, April, 1978, 156 pp. (PDF)Anchor TR-472-report TR-472-report
TR-655. Lemkin, P.F.: The run length map: A representation of contours and regions for efficient search and low level semantic encoding. College Park, MD, University of Maryland Computer Science Center TR-655, April, 1978, 60 pp. (PDF)Anchor TR-655-report TR-655-report
TR-BAS78. Shapiro, B.S.: "Shape description using boundary sequences", U. Maryland Computer Science Dept, 1978. (Ph.D. dissertation).Anchor TR-BAS78-report TR-BAS78-report
...
Peter F. Lemkin, Lemkingroup.com
(Note: You may also access the PDF and journal articles on the lemkingroup.com RTTP history mirror .)
Original: 02/25/2007, Version #57 - original released to History of NIH
Revised: 9/5/2011, Version: #59 - fixed missing links and navigation change
Content transferred from html to confluence: Spring 2019, Fixed footnote page anchors that weren't functioning.